Finding genes associated with disease through multiple-trait-colocalization (MOLOC)
By Robert Smith, Claudia Giambartolomei, James Boocock, and Huwenbo Shi, March 5th, 2018
MOLOC, a method recently developed in our lab by Giambartolomei et al. (A Bayesian Framework for Multiple Trait Colocalization from Summary Association Statistics) integrates GWAS summary data with molecular phenotypes such as gene expression (expression QTL or eQTL) and DNA methylation (methylation QTL or mQTL). Through colocalization, MOLOC pinpoints shared variants across these different data types and identifies those which are responsible for driving association signals. By layering information in this way, this method increases power as well as provides insight into the functional importance of genes.
MOLOC integrates GWAS results for complex phenotypes and is the first colocalization method to use information from three distinct data types. Specifically, in this application, it is used it to link information from methylation and expression and may be applied to any trio of molecular phenotypes.
What is colocalization?
The idea behind colocalization is to identify shared variants across different, but related molecular phenotypes to better understand how these variants affect downstream pathways and suggest those which may be casual for disease. This is done by identifying SNPs at a particular locus which are responsible for two or more molecular phenotypes.
Colocalization methods have been successfully employed to increase power and understand the implications of genetic variation on pathways. They work by identifying shared causal variants behind different associated datasets and are a powerful way of linking genes to disease.
Understanding pathways
Linking molecular phenotypes to genes can help us understand biology. Using MOLOC, we can link a methylation probe to a gene. In our study using this technique, we observed increased CpG methylation in promoter regions that are associated with silencing of gene expression. This is in contrast to findings from genome-wide expression and methylation studies where the correlation between methylation and gene expression observed if often low or the pattern of association is mixed.
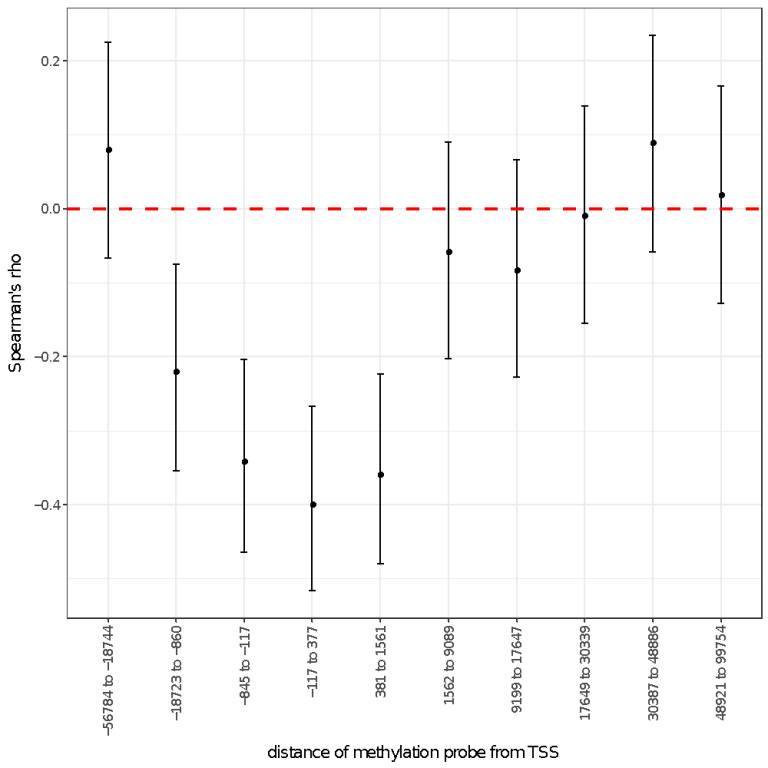
Increasing power
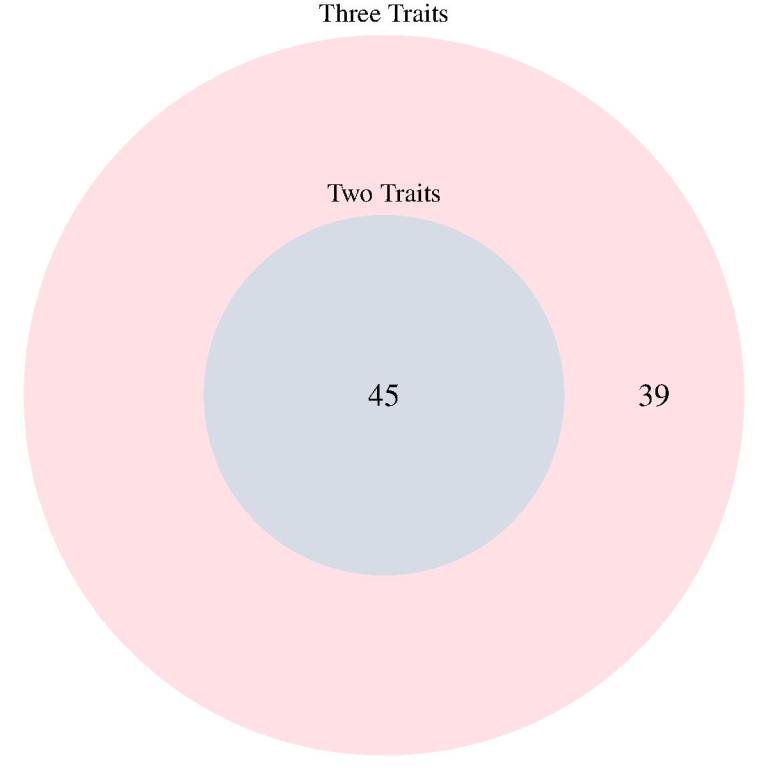
Colocalization of additional layers of molecular phenotypes increases power to find genes related to disease. MOLOC does this particularly well by using three traits.
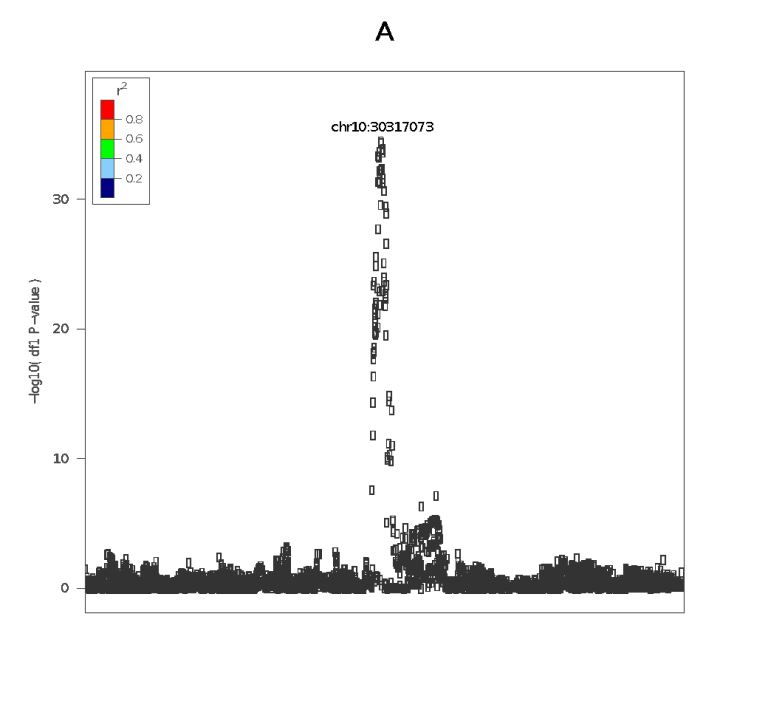
A recent application
We recently applied MOLOC to find shared genetic variation between a precursor of coronary artery disease (carotid intima media thickness – cIMT), gene expression in aorta, and cardiovascular disease. Here, we identified the gene candidate KIAA1462 locus (chrom 10).
New features, recently added to MOLOC (added on 02/15/2018):
- Easy to use for genome-wide data, or any region defined from a bed file.
- Integrate per-SNP functional annotations. This is particularly useful if we would like to link the priors to vary by SNP, for example, to determine if a SNP falls within a particular annotation.
If you would like any further information, or have questions in regards to how MOLOC may be applicable to your project please don’t hesitate to contact Claudia at claudia.giambartolomei@gmail.com
MOLOC is publically available as an R package here: https://github.com/clagiamba/moloc.
