Identifying causal paths from SNP to chromatin to gene expression (Pathfinder)
by Robert Smith, Megan Roytman, March 13th, 2018
A paper recently published in our lab by Roytman et al. "Methods for fine-mapping with chromatin and expression data" presents a fine-mapping framework which computes posterior probabilities for causal paths from SNP to gene expression through chromatin.
The basis of this hypothesis was drawn from knowledge that a significant amount of SNPs in regulatory regions of the genome are simultaneously associated with histone modifications (changes in chromatin) and gene expression.
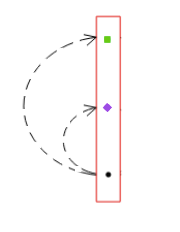
Pathfinder takes as input SNP-chromatin mark and chromatin mark-expression association statistics, as well as correlations on both the SNP and mark levels. Through simulations, this paper demonstrates that Pathfinder has well-calibrated posterior probabilities and its SNP-finding and mark-finding accuracy are higher than existing frameworks that do not take into account associations on both the SNP-mark and mark-expression levels. Pathfinder was applied to every gene in the genome that showed evidence for this flow of causality and the top resulting paths demonstrated consistency with the proposed causal model and often lie in likely functional regions.
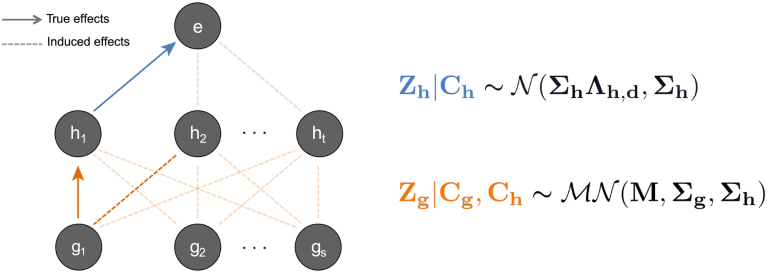
Below is an illustration from the paper of a hypothetical region centered around a gene that Pathfinder may be applied to, accompanied by the mathematical model we use to fine-map SNPs and chromatin marks.
If you would like any further information, or have questions in regards to how Pathfiner may be applied to your project, please don’t hesitate to contact Megan at meganroytman@gmail.com
We regularly post new content, see more below:
On papers:
- Identifying causal paths from SNP to chromatin to gene expression (Pathfinder) by Robert Smith, and Megan Roytman, Mar. 13th, 2018; based on paper: "Methods for fine-mapping with chromatic and expression date"
- Insights from ‘zooming in’ to look at Local Genetics Correlation using Summary Statistics (ρ-HESS) by Robert Smith, Huwenbo Shi, and Nick Mancuso, February 8th, 2018; based on paper: Shi H., Mancuso N., Spendlove S., Pasaniuc B. Local genetic correlation gives insights into the shared genetic architecture of complex traits
- Finding genes associated with disease through multiple-trait-colocalization (MOLOC) by Robert, Claudia, James, and Huwenbo; based on paper: A Bayesian Framework for Multiple Trait Colocalization from Summary Association Statistics
About software:
- Visualizing fine-mapping studies with CANVIS, by Ruth Johnson, January 25th, 2018
Other:
- Tips for Formatting A Lot of GWAS Summary Association Statistics Data, by Huwenbo Shi, February 2nd, 2018
- Explaining Missing Heritability Using Gaussian Process Regression (Reader's Digest), by Huwenbo Shi, Feb. 16th, 2018
